1. INTRODUCTION
ResMarkerDB is a Platform of Biomarkers of response to breast and colorectal cancer monoclonal antibody therapy that will enable the prediction of patients’ clinical outcome after treatment. The focus of our database are FDA approved antibodies for breast and colorectal cancer: anti-EGFR and anti-VEGF antibodies (cetuximab, panitumumab, ramucirumab and bevacizumab), the first two for wild-type KRAS and NRAS colorectal cancer; and anti-HER2 antibodies (trastuzumab, pertuzumab and the antibody-drug conjugate trastuzumab-emtansine) for HER2-positive breast cancer. This information has been collected from available public sources containing data showing the relationship between a biomarker, breast or colorectal cancer, one of the targeted monoclonal antibody treatments and the patient’s response. These resources are: “CGI”, “CIViC”, "JAX-CKB" and “ncDR”. Moreover, we have added data collected by text mining and posterior manual curation. Future improvements encompass the inclusion of more data collected by text mining.
ResMarkerDB aims to integrate the different entities considered with an appropriate terminology. Existing ontologies have helped us in this direction offering specificity to the different entities and the relationships involved. Therefore an effort has been done to standardize all the data employed by the different source databases.
2. DATA SOURCES
ResMarkerDB database is developed by the integration of data from different repositories, "CGI", "CIViC", "JAX-CKB" and "ncDR" as well as from original "ResMarkerDB- ResCur" data obtained by text mining process.
- “CGI” or “Cancer Genome Interpreter” aims to detect therapeutically actionable tumor alterations driving cancer among all the identified alterations. It provides a cancer biomarkers database which integrates manually collected genomic biomarkers of drug response associated with a tumor type and level of evidence. Data was downloaded on 2018/07/16. (Tamborero et al., 2017)
- “CIViC” or “Clinical interpretations from Variants in Cancer” is an open resource for Clinical interpretation of Variants in Cancer which tries to enable precision medicine. Data was downloaded on 2018/07/16. (Griffith et al., 2017)
- "JAX-CKB” or “The Jackson Laboratory Clinical Knowledgebase (CKB)” is a curated database of gene/variant annotations, therapy knowledge, diagnostic/prognostic information, and clinical trials related to oncology. Data was downloaded on 2018/07/03. (Patterson et al., 2016)
- "ResCur" referred source is used for those evidences original from our database. These data were obtained by text mining process conducted by means of Pubtator and SCAIView, which are text mining tools available online. Both of them were used to annotate different entities in order to complement each other. After that a manual curation process was made. Moreover, we collected data from “ncDR” or “ncRNA in Drug Resistance” (Dai et al., 2017), a database resource that collects non-coding RNA linked with its drug-response to understand the underlying molecular mechanisms of drug response. Then, these data were further characterized using Pubtator to specify the ncRNA alteration and annotate it with a higher level of granularity. Again, this data was manually curated by an expert. We also refer to this dataset as "ResCur".
3. DATABASE CONTENTS
ResMarkerDB provides biomarkers associated to drug response in specific tumor types with their supporting evidence. Specific information on drug response, evidence level, original data source and an example statement are provided too. Moreover, a process of standardization and homogenization of the information collected from the different sources was performed.
3.1. Genes
Genes are referred with NCBI Entrez Gene® identifiers.
3.2. Biomarkers
In ResMarkerDB we include drug response biomarkers defined as genetic or epigenetic alteration objectively measured which serves as indicator of the treatment response in a certain cancer type according to distinct levels of evidence. We include the following types of drug response biomarkers:
- Gene variants: includes different types of alteration in the gene sequence related to the drug response. Covers the following types described in the original source databases: wild-type, variants, mutation, oncogenic mutation, exon mutation, codon mutation, bialellic inactivation, truncations, and small insertions and deletions.
- Copy number alterations: includes different types of alteration in the copy number of the gene sequence related to the drug response. Covers the following types described in the source databases: amplification and deletion.
- Expression: includes different types of alterations in the expression of the gene or protein related to the drug response. Covers the following types described in the source databases: expression, overexpression, underexpression and positive.
- Functional events: describes functional events of the protein related to the drug response. Currently it only includes the event nuclear translocation reported by “CIViC”.
3.3. Drugs
ResMarkerDB focuses on FDA-approved monoclonal antibodies for the target cancer types, including drug combinations. Combination therapies included are combinations of a monoclonal antibody therapy with an antineoplastic and immunomodulating agents and/or another monoclonal antibody therapy.
Drugs for Breast Cancer:
- Trastuzumab: anti-HER2 antibody for HER2 positive breast cancer.
- Pertuzumab: anti-HER2 antibody for HER2 positive breast cancer.
- Trastuzumab-emtansine: anti-HER2 antibody-drug conjugate.
- Cetuximab: anti-EGFR antibody for wild-type KRAS and NRAS colorectal cancer.
- Panitumumab: anti-EGFR antibody for wild-type KRAS and NRAS colorectal cancer.
- Bevacizumab: anti-VEGF antibody.
- Ramucirumab: anti-VEGF antibody.
3.4. Tumor Types
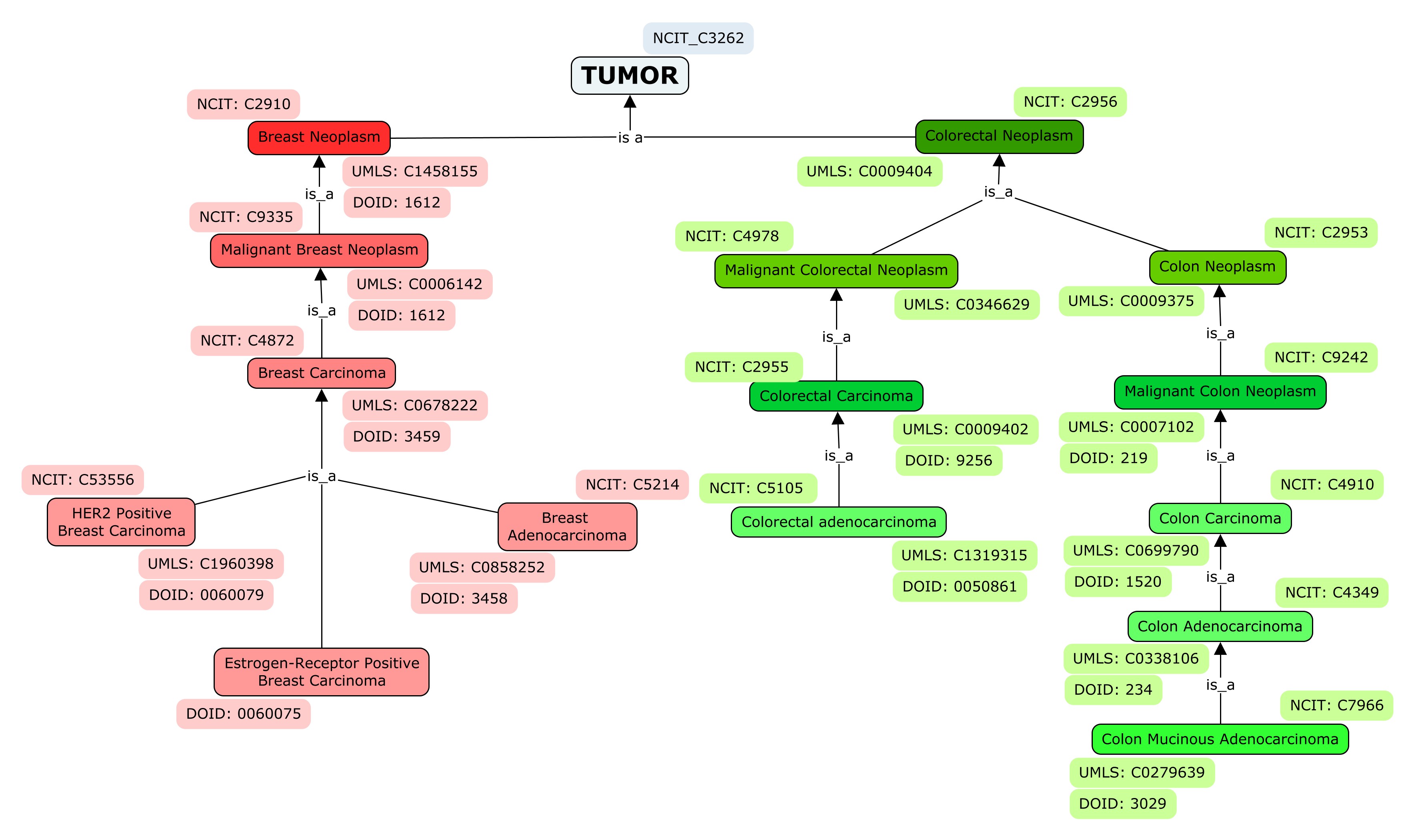
ResMarkerDB focuses on malignant breast and colorectal cancer. The different cancer subtypes covered are briefly described below, with reference to the original terms used by the source databases. To ease the comprehension, in this documentation we will refer to the most general groups as breast or colorectal cancer.
The terminology used for tumor types is the NCI Thesaurus OBO Edition® (NICT®), Unified Medical Language System® (UMLS®) and the Disease Ontology® (DOID®).
The terms are hierarchically organized according to NCIT® and also cross-referenced to DOID® and UMLS® identifiers. Moreover, synonymous terms extracted from NCIT® are included in our database to ease the web searches.
- Breast Neoplasm: benign or malignant neoplasm of the breast parenchyma.
- Malignant Breast Neoplasm: a primary or metastatic malignant neoplasm involving the breast.
- Breast Carcinoma: abnormal proliferation of cells deriving from epithelial cells of the breast. Tries to cover the term breast cancer used by the origin databases “CGI”, “CIViC”, “JAX-CKB” and “ncDR”.
- Breast Adenocarcinoma: it has origin in the milk ducts and/or lobules (glandular tissue) of the breast.
- Inflammatory Breast carcinoma characterized by being invasive and by the presence of different changes in the overlying skin. The same exact term is used by “CIViC”.
- HER2 Positive Breast Carcinoma: biologic subset of breast carcinoma defined by high expression of HER2, GRB7 and TRAP100, and by lack of expression of estrogenic receptor. It stands for Her2-receptor positive breast cancer from “CIViC” and “JAX-CKB”.
- Estrogen-Receptor positive Breast Carcinoma: biologic subset of breast carcinoma characterized by the presence of the receptor protein that binds estrogen, which may be needed to grow. It comes from “JAX-CKB”.
- Breast Adenocarcinoma: it has origin in the milk ducts and/or lobules (glandular tissue) of the breast.
- Breast Carcinoma: abnormal proliferation of cells deriving from epithelial cells of the breast. Tries to cover the term breast cancer used by the origin databases “CGI”, “CIViC”, “JAX-CKB” and “ncDR”.
- Malignant Breast Neoplasm: a primary or metastatic malignant neoplasm involving the breast.
- Colorectal Neoplasm: benign or malignant large intestine neoplasm affecting the colon and/or rectum. Tries to encompass colorectal cancer coming from “CGI”, “CIViC”, "JAX-CKB" and “ncDR”.
- Malignant Colorectal Cancer: a primary or metastatic malignant neoplasm that affects the colon or rectum.
- Colorectal Carcinoma: a malignant epithelial neoplasm that arises from the colon or rectum and invades through the muscularis mucosa into the submucosa.
- Colorectal Adenocarcinoma: characterized specifically by malignant epithelial cells of glandular origin invading through the muscularis mucosa into the submucosa. It covers exact term from “CIViC” and “JAX-CKB”.
- Colorectal Carcinoma: a malignant epithelial neoplasm that arises from the colon or rectum and invades through the muscularis mucosa into the submucosa.
- Colon Neoplasm: benign or malignant neoplasm specifically located in the colon.
- Malignant Colon Neoplasm: primary or metastatic malignant neoplasm that affects the colon.
- Colon Carcinoma: abnormal proliferation of cells deriving from epithelial cells a rising from the colon and invades through the muscularis mucosa into the submucosa. The term tries to encompass colon cancer referenced in “CIViC” , “JAX-CKB” and “ncDR”.
- Colon Adenocarcinoma: it derives from epithelial cells of glandular origin.
- Colon Mucinous Adenocarcinoma: characterized by the presence of pools of extracellular mucin. the exact term appears in “CIViC”.
- Colon Adenocarcinoma: it derives from epithelial cells of glandular origin.
- Colon Carcinoma: abnormal proliferation of cells deriving from epithelial cells a rising from the colon and invades through the muscularis mucosa into the submucosa. The term tries to encompass colon cancer referenced in “CIViC” , “JAX-CKB” and “ncDR”.
- Malignant Colon Neoplasm: primary or metastatic malignant neoplasm that affects the colon.
- Malignant Colorectal Cancer: a primary or metastatic malignant neoplasm that affects the colon or rectum.
3.5. Response
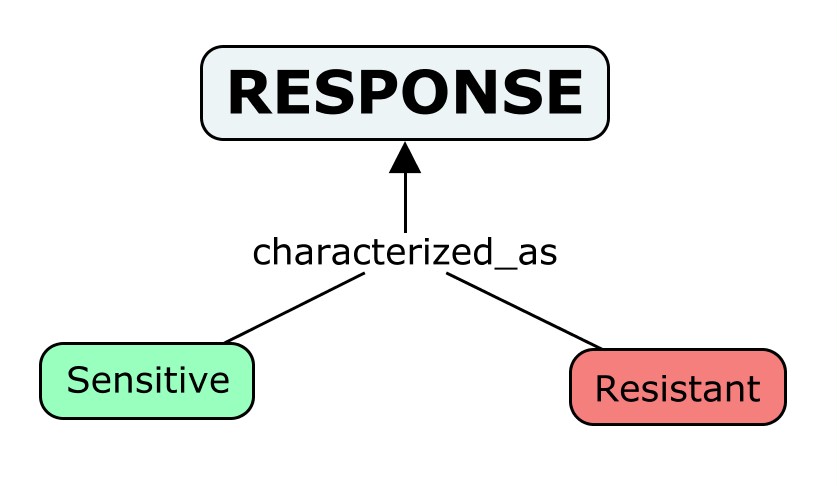
In ResMarkerDB we consider the NCIT definition (NCIT_C50995) of Response as “the pathologic and/or clinical changes that result from treatment. The changes may include eradication of detectable disease, stabilization of disease, or disease progression”.
Under this concept we integrate association used in “CGI”, clinical significance used by “CIViC”, response_type used by “JAX-CKB” and effect used by ncDR. It can be sub-classified the following way:
- Sensitive: response to a drug used to kill or weaken cancer cells. It includes responsive from “CGI”, sensitivity from "CIViC" and sensitive from “JAX-CKB” and “ncDR”.
- Resistant: failure of cancer cells to respond to a drug used to kill or weaken them; that is being in a state or condition of lack or negative response to a pharmacological agent. Those cells may be resistant to the drug at the beginning of the treatment, or may become resistant after being exposed to it. It includes the term no-responsive from “CGI”; resistance or non-response from “CIViC”; resistant from “CGI”, “JAX-CKB” and “ncDR”; and no benefit from “JAX-CKB”.
3.6. Evidence level
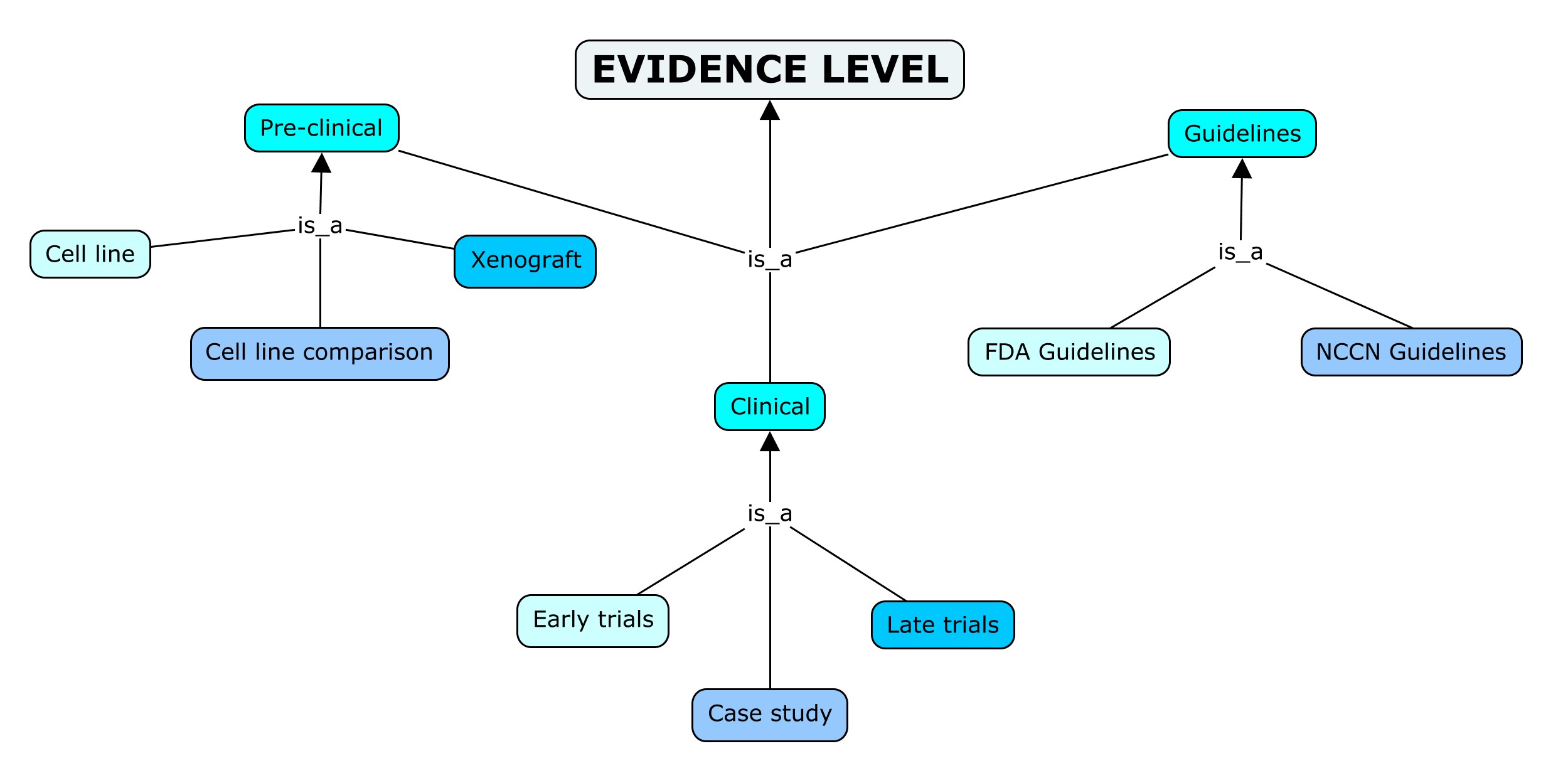
In ResMarkerDB we use the NCI definition (NCIT_C15639) of Evidence Level: “A formal ranking system of the strength of evidence linked to a reported result”. Under this concept we integrate the terms evidence level of “CGI” and “CIViC”, approval status of “JAX-CKB” and Materials 1-4 of “ncDR”. We provide three main types of evidence (preclinical, clinical and guidelines) as described below:
- Preclinical: originally coming from research done in a laboratory that may use special equipment and cells or animals using in vivo or in vitro models. Used the same way in “CGI”, “CIViC” and “JAX-CKB”, and it covers materials 1, 2 and 3 from “ncDR”.
- Cell line: permanently established cell culture that will proliferate indefinitely given appropriate fresh medium and space. Covers cell line and cell culture by “JAX-CKB” and what is known as materials 1 in “ncDR”.
- Cell line comparison: contrast between different cell lines; referred as materials 2 in “ncDR”.
- Xenograft: transfer cells, tissues, or organs from a donor into another species. Considers xenograft and patient derived xenograft (Pdx) used by “JAX-CKB” and materials 3 in “ncDR”.
- Clinical: research conducted in human subjects. The term is employed by “CIViC”. This category includes in a subdivision the case report term from “CGI” and case study used by “CIViC” and “JAX-CKB”. Moreover, it encompasses the terms early and late trials used by “CGI”; different phases referred in “JAX-CKB”; and materials 4 from “ncDR”.
- Early Trials: clinical trials I-II. Encompasses the terms Phase I, Phase Ib/II and Phase II employed by “JAX-CKB”.
- Late Trials: clinical trials III-IV, that is therapeutic and prognostic or diagnostic studies. Includes the term Phase III used by “JAX-CKB”.
- Case Study: uncontrolled observational study reporting an individual case from clinical journals, detailing the diagnosis, treatment, and follow-up of an individual patient.
- Guidelines: systematic statement stating a proven or consensus association in human medicine. These are guidance or protocols developed to help health care professionals and patient make decisions about treatment of a specific health condition in our case. Originally it is referred as Validated by “CIViC”.
- NCCN guidelines: from NCCN (“National Comprehensive Cancer Network”). Stated the same way by “CGI”.
- FDA Guidelines: from FDA (“Food and Drug Administration”). Referenced as FDA Guidelines by “CGI” and FDA approved by “JAX-CKB”.
3.7. Reference
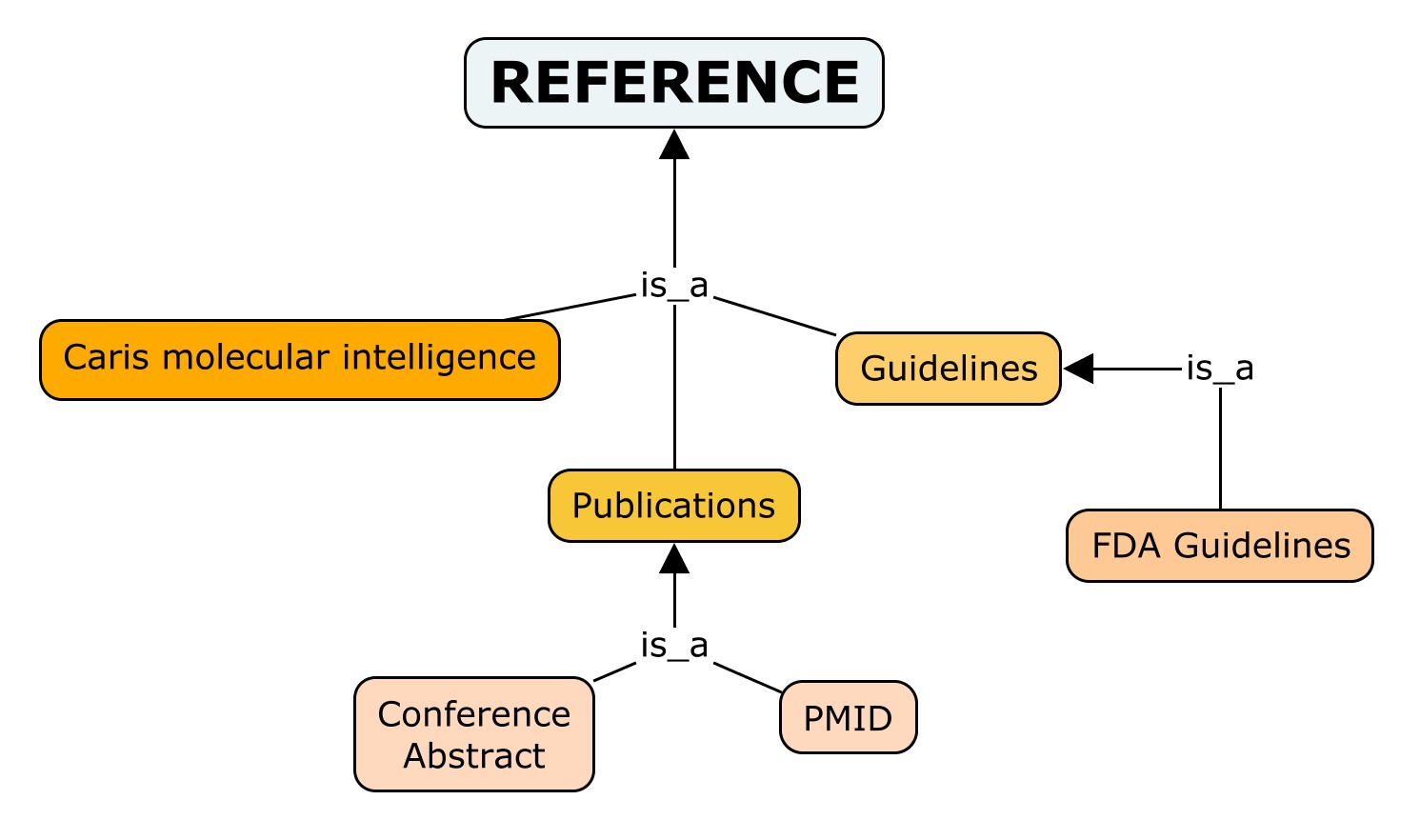
As Reference we provide the reference supporting the association where data come from. It covers the source or reference from “CGI”, PubMed ID from “CIViC”, references in “JAX-CKB” and literature info (PMID and Support) from “ncDR”. The references can be divided the following way:
- Guidelines: systematic statement of policy rules or principles, international clinical recommendations. Used by "JAX-CKB".
- FDA Guidelines: guidelines developed by the “Food and Drug Administration”, from “CGI”.
- Publications: printed or electronic work offered for distribution covering the association evidenced.
- Conference Abstracts: brief summary of a paper presented in a conference, from “CGI” and “JAX-CKB”.
- PMID: PubMed ID for the publication where the association was described, from all databases.
- Caris molecular Intelligence: Science Company at the forefront of precision medicine, from “CGI”.
3.8. Statement
In ResMarkerDB we provide a Statement asserting or declaring the evidence association. It is reported by “CIViC” as evidence statement or description, by means of a summary of the evidence’s potential clinical interpretations; efficacy evidence by “JAX-CKB”, and as support (a sentence extracted from the publication) by “ncDR”.
4. STATISTICS
4.1. Genes
Classification of genes using PANTHER Classification System.
- Molecular function
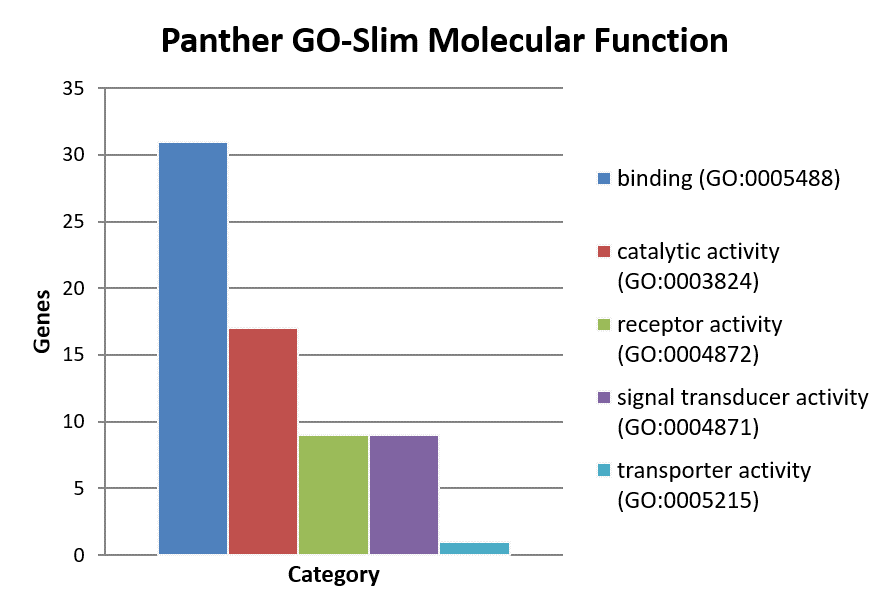
 ××
×× - Biological Process
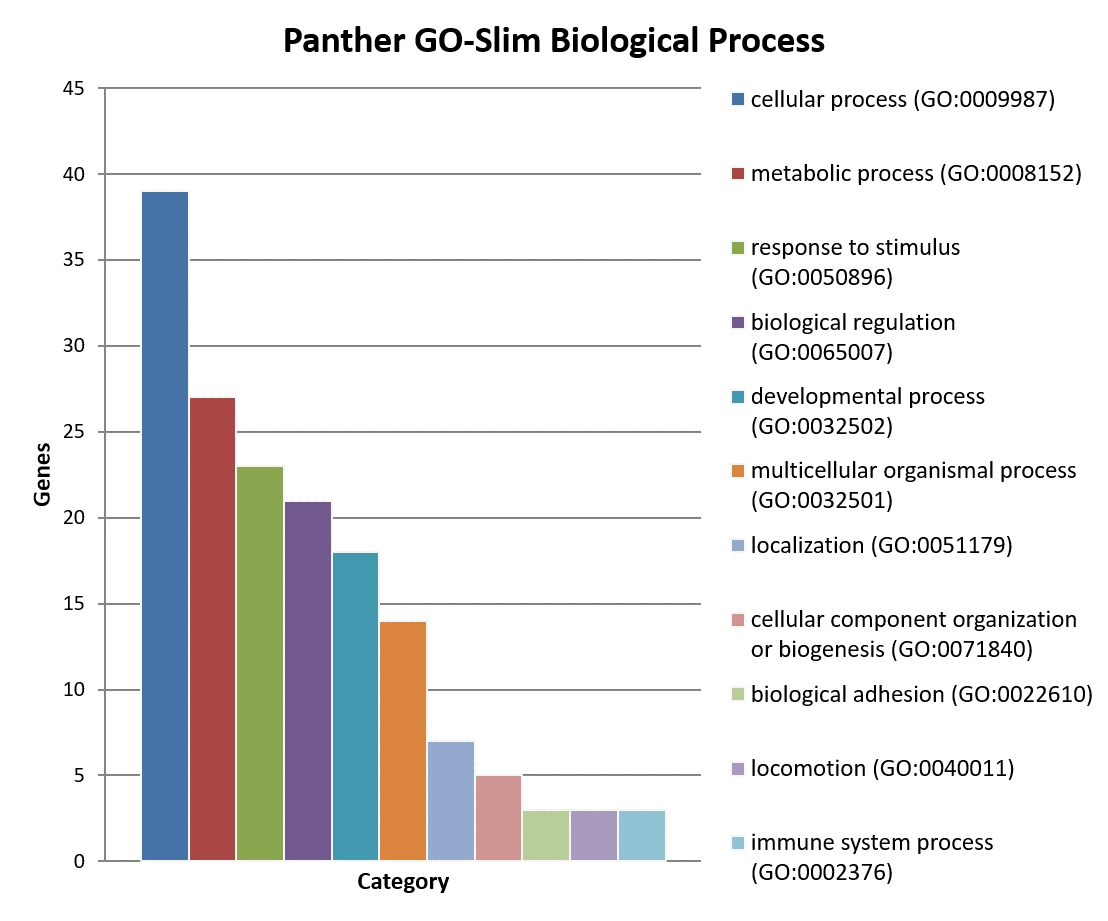
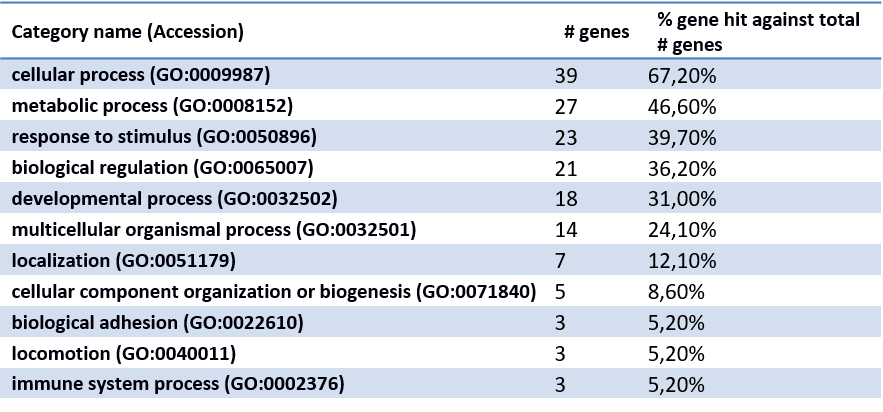 ××
××
4.2. Biomarkers
| Biomarkers | Biomarkers per Database |
|---|---|
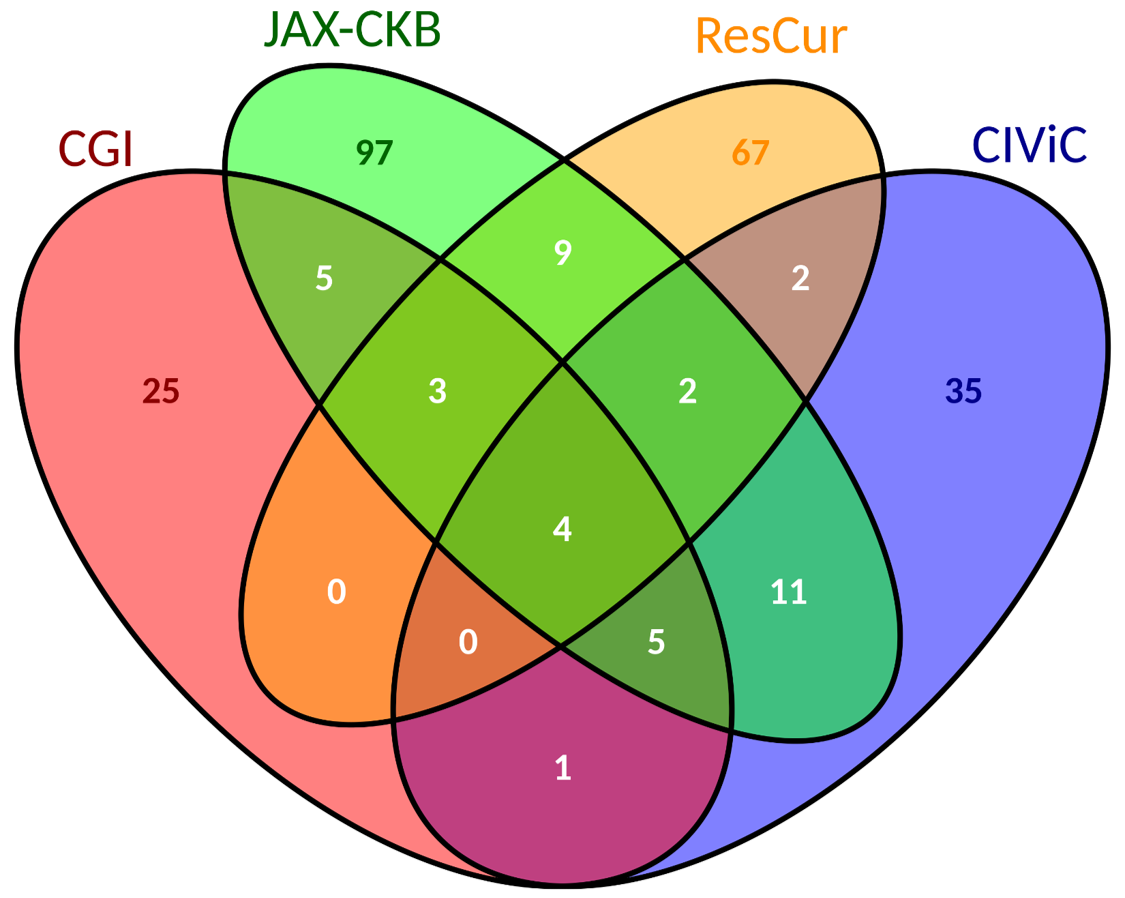
| There are a total of 266 biomarkers, that include gene variants, copy number alterations, etc. We have a total of 28 combinations of variants and other kinds of biomarkers. Miscellaneous category covers: Copy Number Alterations, Expression Alterations, Functional Events, Protein Serum Levels or combinations of them. | There are 9 biomarkers shared by the databases "CGI", "CIViC" and "JAX-CKB", 4 of which are also shared with "ResMarkerDB". |
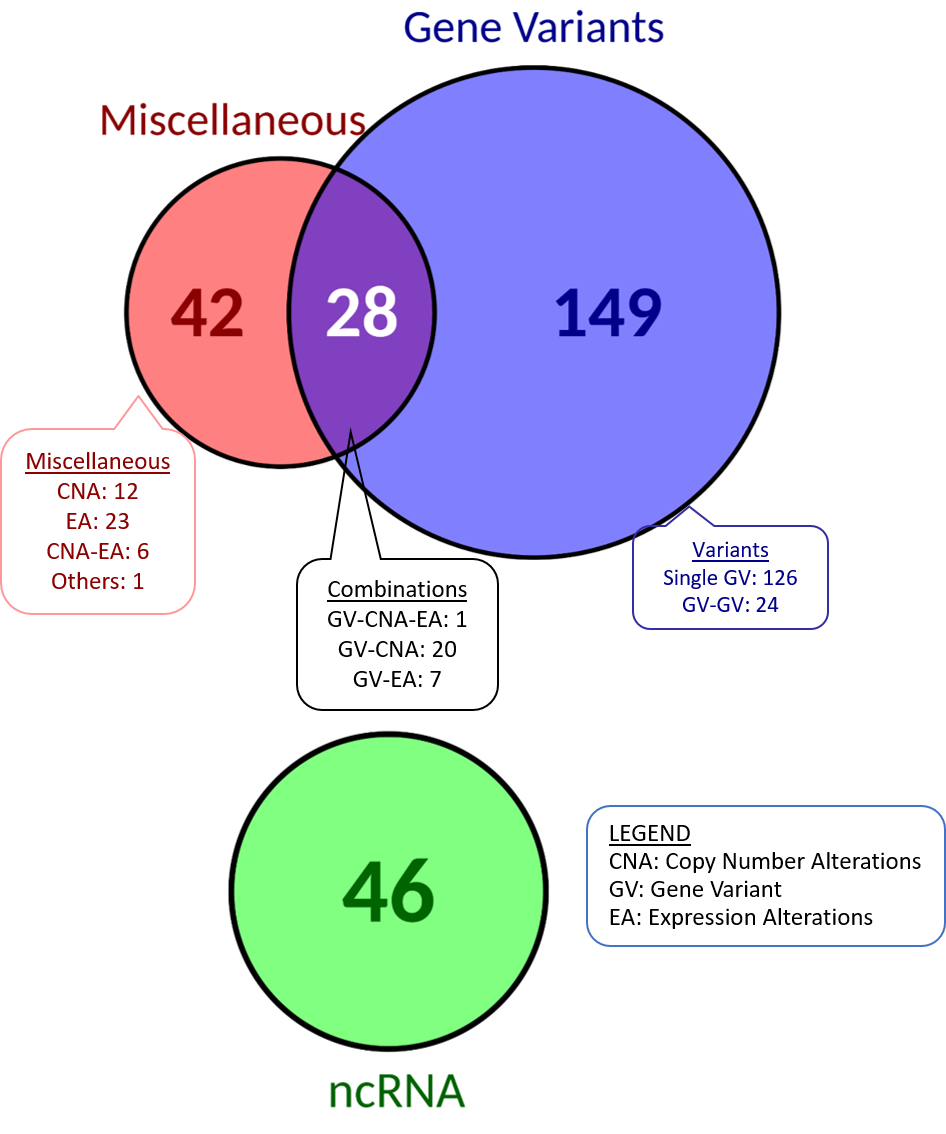
×
|
×
|
4.3. Drugs
| Drugs per Database | Drugs Classification |
|---|---|
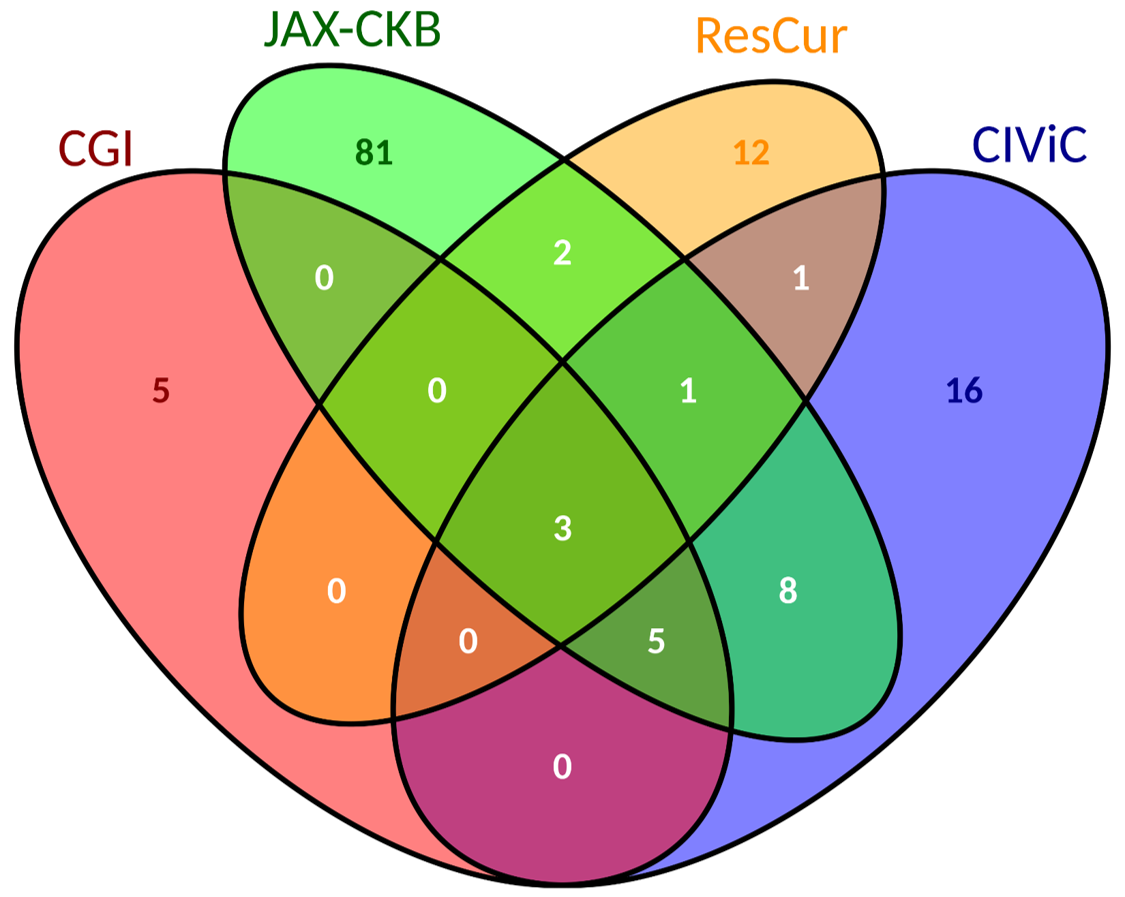
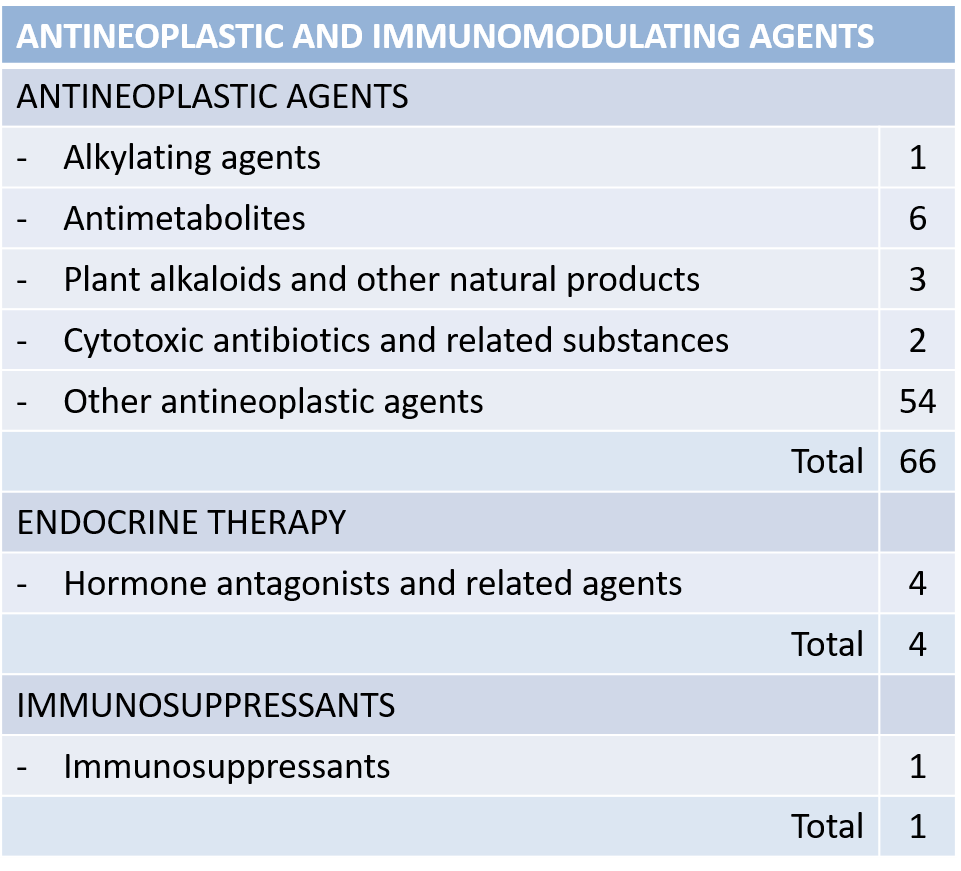
| There is a total of 134 reported treatments and 73 drugs. Of them, only 3 are shared among all sources: "Trastuzumab", "Cetuximab" and "Panitumumab". | Anatomical Therapeutic Chemical (ATC) Classification from WHO. |
×
|
×
|
4.4. Tumor Types
| Biomarkers per Tumor type | Breast Cancer Biomarkers per Source | Colorectal Cancer Biomarkers per Source |
|---|---|---|
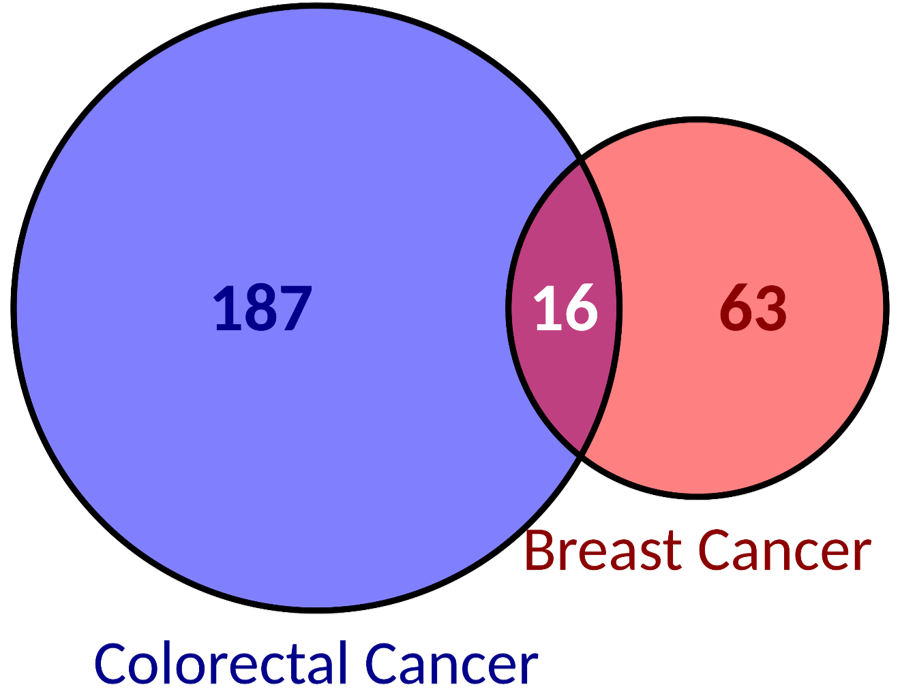
| We report more biomarkers associated to colorectal cancer than to breast cancer. | Source of biomarkers associated to breast cancer. | Source of biomarkers associated to colorectal cancer. |
×
|
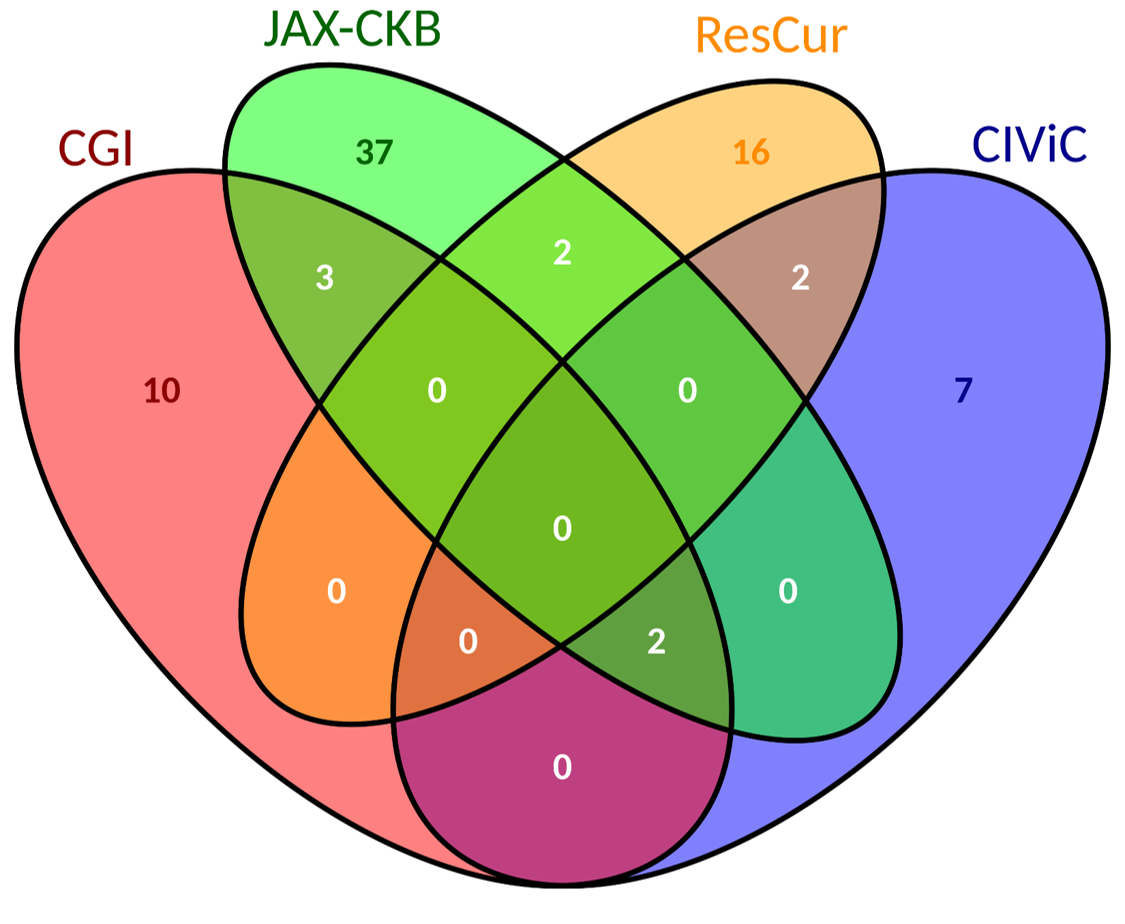
×
|
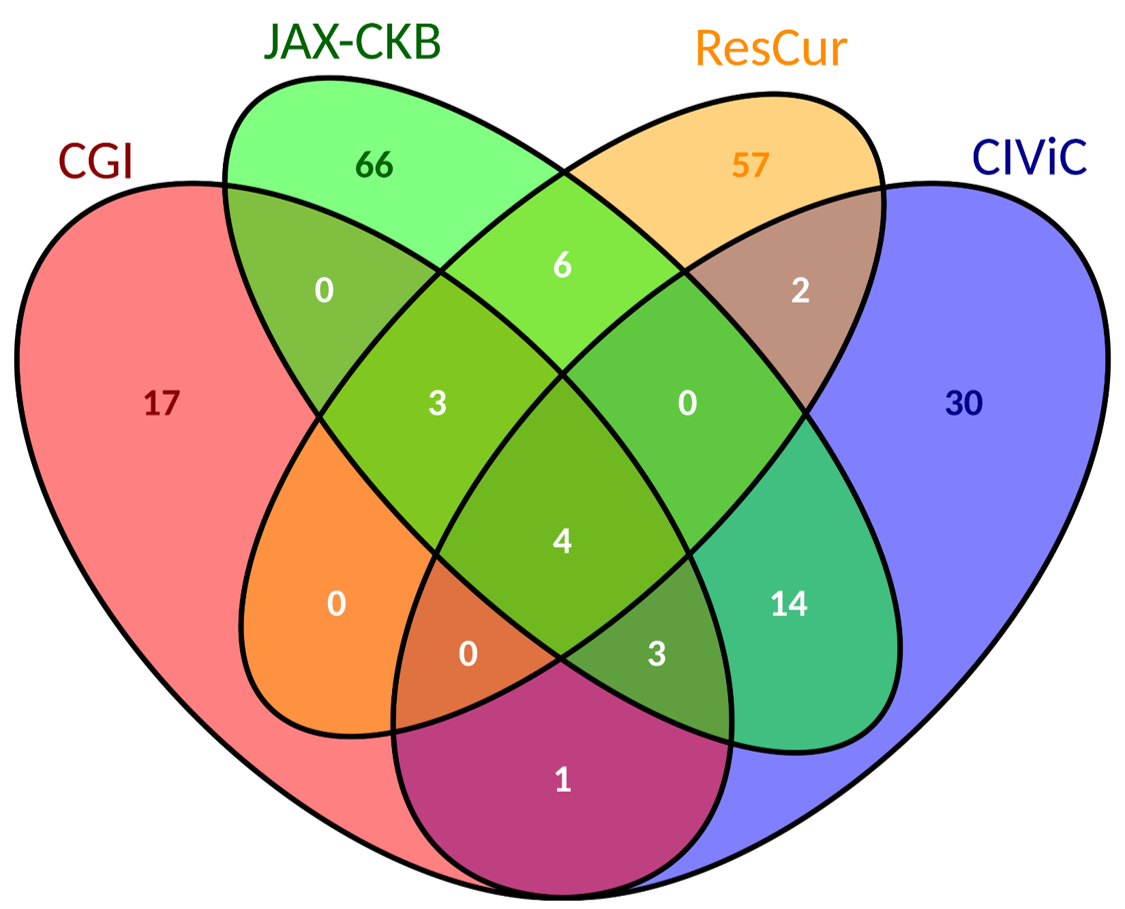
×
|
4.5. Response
| Biomarker-Drug-Tumor trios per Response |
|---|
| There are approximately the same number of biomarker-drug-tumor combinations associated to resistance and to sensitivity. Moreover, there are three of those combinations that have opposite responses. This can be partially explained by the evidence level. Interestingly those biomarkers may show sensitivity to treatment at a preclinical level but resistance at a clinical level. |
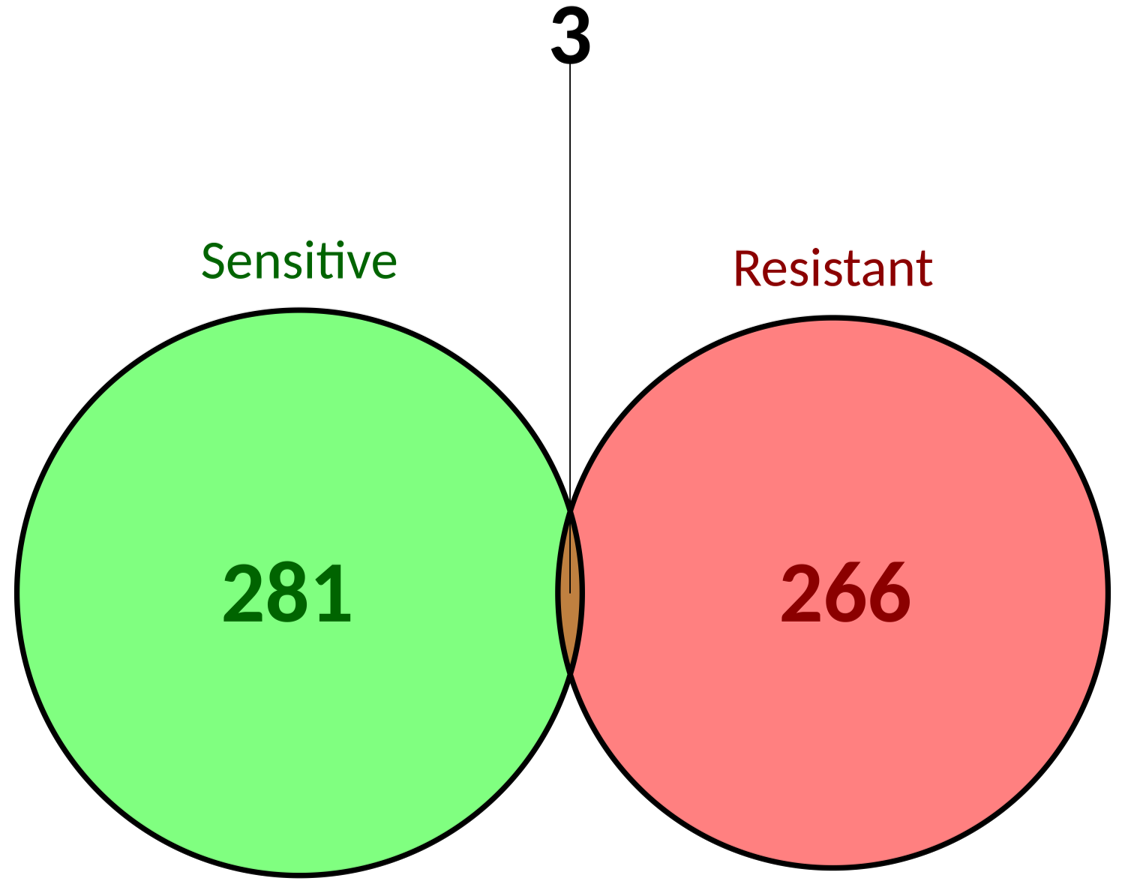
×
|
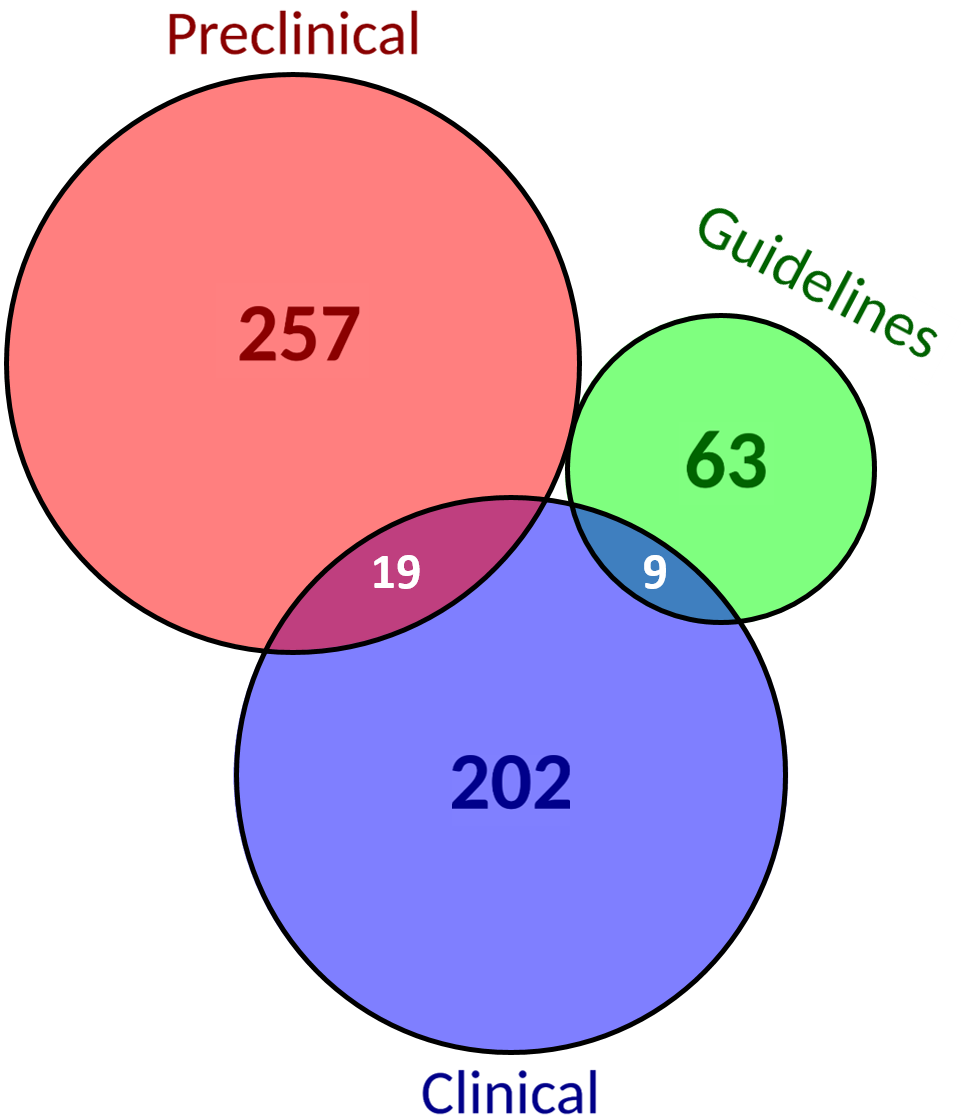
4.6. Evidence Level
| Biomarker per Evidence | Biomarker-Drug-Tumor trios per Evidence |
|---|---|
| 55% of biomarkers are reported at preclinical levels and 51% at clinical levels. | There are no biomarker-drug-tumor combinations reported in all evidence levels considered. The percentage of those combinations evidenced at the different evidence levels is almost kept the same as when only considering biomarkers. |
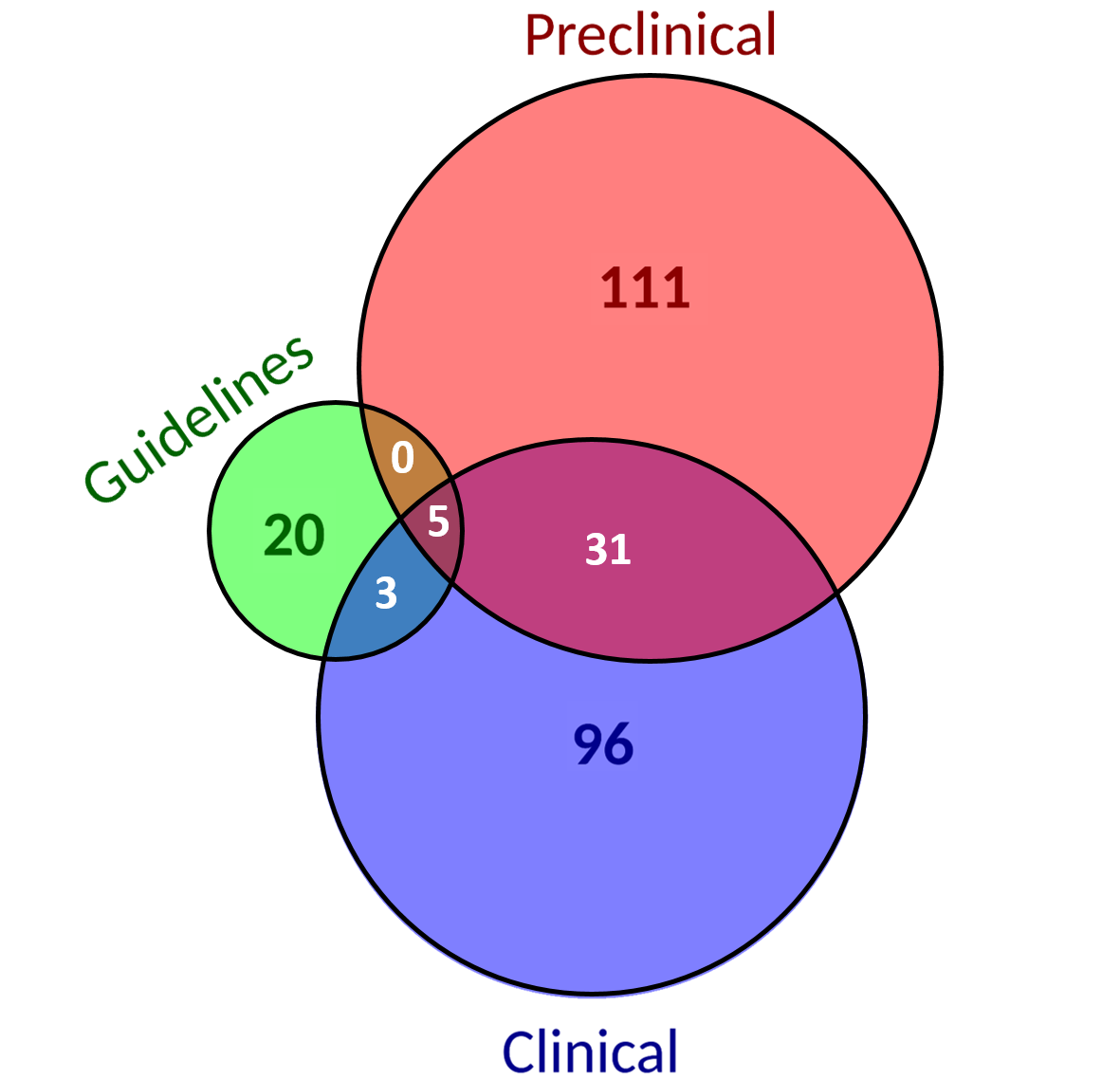
×
|
×
|
5. CHALLENGE DATA
ResMarkerDB has also proven to be useful in the field of precision medicine. The same database structure with amplified contents has been used in the context of the '2018 TREC Precision Medicine / Clinical Decision Support Track’. The main aim was to support the retrieval of article reporting tailored treatments for cancer patients with specific genomic alterations by a text mining process. We have made part of this dataset publicly available under CC BY 4.0.
2018 TREC Precision Medicine Challenge Data